PDB Structures & ONIOM
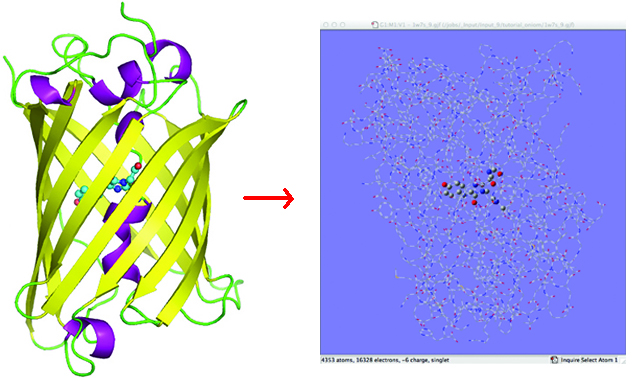
The ONIOM method is powerful tool for modeling biological systems. Such compounds typically consisting of a large protein environment as well as a region of interest. The latter may be another molecule bonded to the protein (e.g., a chromophore), an active site within the protein, or something else.
One challenge with using the ONIOM method for such problems is preparing appropriate input for the calculation. Once set up, ONIOM jobs are not fundamentally different from other calculations. However, preparing a molecule specification for use with the ONIOM method in Gaussian is a rather lengthy process. While not excessively difficult, it is nevertheless challenging at times, and
each part of it requires patience and care.
In general, creating a Gaussian input file for a biological molecule encompasses the following steps:
- Obtaining a starting PDB file.
- Adding hydrogen atoms to and specifying protonation for the PDB file structure.
- Specifying parameters for the molecular mechanics method (e.g., Amber): atom types and partial charges.
- Assigning atoms to ONIOM layers and defining link atoms.
- Selecting model chemistries for each ONIOM layer.
- Running the desired calculation(s).
The initial part of chapter 9 discussed this process in considerable detail. It includes discussion of the variety of software packages that can be useful in the process.






